COVID-19
SARS-CoV-2 Proteins
The SARS-CoV-2 genome is known to encode at least 29 proteins, all of which are of great interest to researchers investigating the virus life cycle, host cell entry, and immune system stimulation, and all for the purpose of seeking out targets for treatments and vaccines.
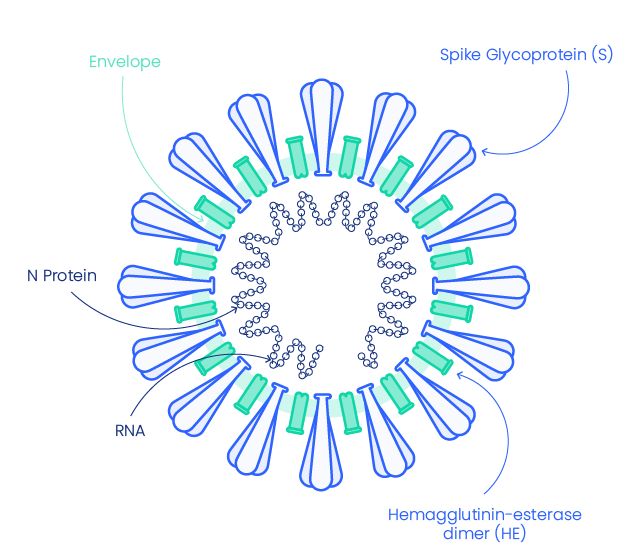
SARS-CoV-2 structure and components
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, 2019-nCoV), the coronavirus responsible for coronavirus disease 2019 (COVID-19), shares very high similarity with SARS-CoV. It appears with a crown-like surface projection on microscopic imaging indicating it belongs to the beta-coronavirus family (1), which are enveloped, single-stranded RNA viruses.
They mainly infect host lung cells through binding to the Angiotensin Converting Enzyme 2 (ACE2) receptor.
SARS-COV-2 Structural Proteins
Structural proteins are essential in many steps of the infection, they are implied in viral genome production, replication, virion-receptor attachment, viron and viroporin formation that will promote virus entry into the host, proliferation and spread of the infection. (2)
Spike glycoprotein
The envelope of corona-virion contains protruding projection from its surface called spike proteins (or S proteins).
The Spike glycoprotein mediates the virus attachment to host cell surface receptors ACE2 and facilitates virus entry by assisting fusion between viral and host cell membranes. It is the most exposed and immunogenic viral protein and hence a target of choice for diagnostic and therapeutic assays.
The Spike glycoprotein of SARS-CoV-2 is a trimeric macromolecule with two furin-like protease cleavage sites. One of the sites is at the boundary between S1 and S2 subunits having poly-basic residues, which is characteristic of SARS-CoV-2. The other cleavage site is located within the S2 subunit.
SARS-CoV-2 Spike Structure
A. Spike proteins form trimers, each consisting of a short intracellular C fragment, a transmembrane moiety and an ectodomain element. The ectodomain is constituted of two subunits S1 and S2. On the S1 subunit, there is a receptor-binding domain (RBD) that recognizes and binds ACE2.
B. SP: signal peptide; NTD: N-terminal domain; RBD: receptor binding domain; RBM: receptor binding motif; FP: fusion peptide; HR1: heptad repeat 1; HR2: heptad repeat 2; TM: transmembrane domain; CD: cytoplasmic domain. The S1/S2 cleavage site is indicated.
Nucleocapsid (N)
N protein is express in the early stage of infection and is the most abundant protein It forms a core of a ribonucleoprotein by binding to viral RNA. It helps RNA to enter in the cell and to interact with cellular components. (3)
N protein has diverse roles including:
SARS-CoV-2 nucleocapsid protein structure
A. N protein forms an “X” shape due to the CTD domain. It is rich in helix and hydrophobe residues that favor binding to nucleic acid and lead to the neutralization of the charges (2)
B. NTD: N-terminal domain; SRL: (SR)-Rich Linker; CTD: C-Terminal domain.
Envelope protein (E)
SARS-CoV-2 Envelop protein (E-protein) is conserved across β-coronaviruses. It is a tiny integral membrane protein that pentamerize to form ionic pore across the membrane called viroporins. It is essential for viral assembly and release.(2)
SARS-CoV-2 Life Cycle
Learn more about the coronavirus life cycle and discover products
to help you in your research.
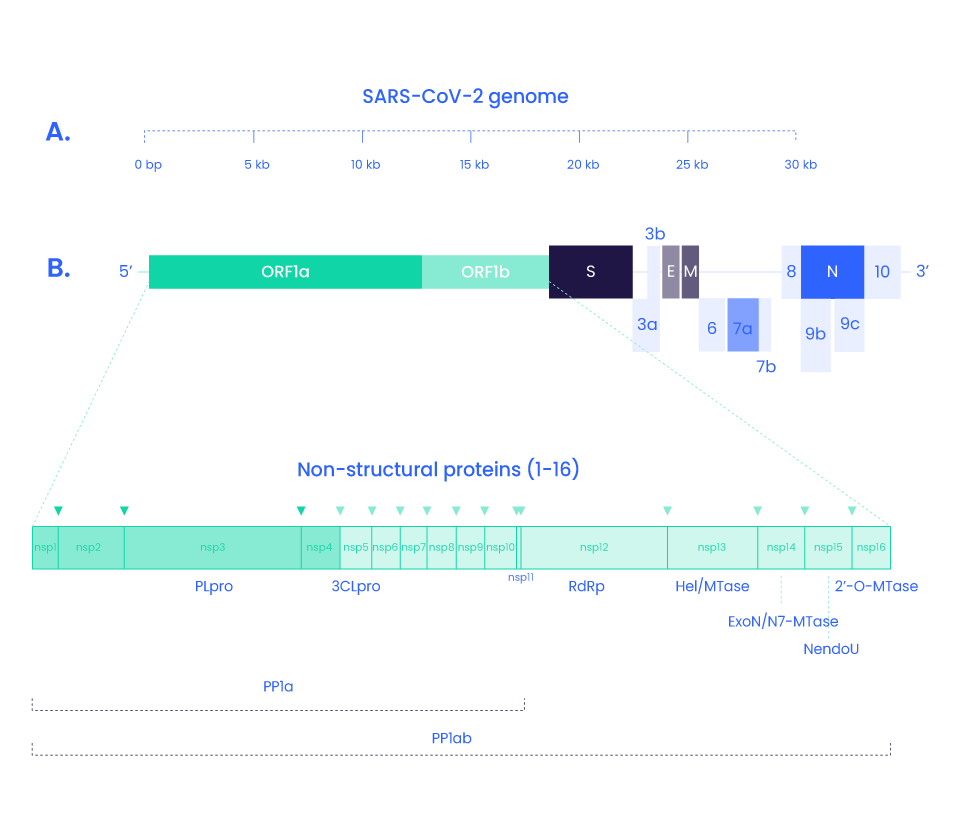
SARS-COV-2 Non-Structural Proteins and peptides
In addition to the four structural proteins, the SARS-CoV2 genome encodes 16 non-structural proteins (NSPs) essential for virus replication but also to elicit the immune response and represent targets to develop future prophylactic and therapeutic approaches against COVID-19 (9). Replication and transcription of the coronavirus are done by a protein complex called RTC (replication/transcription complex)
Some data about protein function in SARS-Cov-2 are not yet known but their homology to their homolog in other Coronaviruses suggests their function might be the same.
3C-like protease is the principal protease of SARS-CoV2 and is essential for RNA replication. It cleaves the polyprotein Orfa with Nsp3 (14). It also cleaves the C-terminus from Nsp4 to Nsp16.
This complex gives rise to the RNA polymerase complex performing de novo initiation and primers extensions to trigger RNA synthesis. (15,16)
Nsp11
Is a short peptide (13 aa) overlapping Nsp10 but its function is still unknown.
Nsp13
NSP13 is the virus helicase, allowing the duplex RNA to unwind and being accessible.(15)
Nsp14
Nsp14 is an exoribonuclease comprised in the RTC complex. It is implied in proofreading and recombination.
Nsp15 (NendoU)
NendoU (Nidoviral RNA uridylate‐specific endoribonuclease) is an endonuclease comprised in the RTC complex.
SARS-COV-2 Accessory Proteins and Peptides
The end of the SARS-CoV genome encodes for 9 additional proteins called Accessory proteins.
|
Orf3a |
This protein is responsible for channel formation. It is essential for infectivity, virulence, and virus release (19) |
|---|---|
| Orf3b |
It’s a small peptide of 22 aa overlapping with Orfa and is a potential IFN-1 antagonist (20) |
| Orf6 |
In SARS-CoV, Orf6 is an IFN antagonist. It has been shown to disrupt transportation of transcriptions factors (e.g. STAT1) (21,22) |
| Orf7a |
In SARS-CoV ORF7a blocks glycosylation of BST-2 by its binding to this growth factor known to interact with IFN. (23) |
|
Orf7b |
This peptide of 43 aa overlaps the sequence of ORF7a. (24) |
|
Orf7b/Orf8 |
A deletion in ORF7b and ORF8 leads to a fusion protein that has been identified in various regions and leads to deletion of 328 nucleotides that represents the transcriptional regulatory sequence thereby increasing N gene transcription. This deletion leads to virus attenuation and reduced replication yet leads to immune evasion. (25) |
|
Orf8 |
Orf8 is a less conserved protein compared to SARS-CoV (only 30% homology). It is implied in recognition by the immune system and transcription of N protein. (25) |
|
Orf9b |
It interacts with mitochondrial import receptor Tom70 resulting in the activation of IRF-3. (14) |
|
Orf9c |
Orf9C had been shown to interact with proteins and modulates the NF-κB pathway and IkB kinases. (14) |
|
Orf10 |
This 38 aa peptide seems unique to SARS-CoV-2, it has no homolog in SARS-CoV. (26). Its function is not yet determined but it doesn’t seem to be essential (27) |
Choose your tools
Choose the products that will help you in your COVID-19 research and drug discovery
We offer custom peptide synthesis and off the shelf peptides for immediate shipment. Our peptide experts has synthesized coronavirus-derived peptides such as critical peptide domains/region or peptide substrates to better understand fusion mechanisms of the viral partical with the host cell membrane.
We also provide NF-KB inhibitors to study the impact of the SARS-CoV2 virus on this pathway. We also offer custom manufacturing of Critical Raw Material (CRM) peptides for demanding applications.
| Catalog peptides | Learn more |
|---|---|
| Custom peptide synthesis | Learn more |
We provide library and peptide sets from SARS-CoV-2 Spike protein for screening applications such as compound testing.
The SARS-CoV viral proteins have been identified as targets of several host proteases, among which Furin, 3CLpro (3C-like viral protease) and Cathepsins (B, L) play roles. Based on this information we offer you access to a series of protease assay kits related to the SARS-CoV-2 mechanism of action.
References
1) Transmission electron microscopy imaging of SARS-CoV-2.
Prasad S, Potdar V, Cherian S, Abraham P, Basu A; ICMR-NIV NIC Team
Indian J Med Res. 2020 Feb & Mar;151(2 & 3):241-243 - DOI: 10.4103/ijmr.IJMR_577_20
2) Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2.
Satarker S, Nampoothiri M.
Arch Med Res. 2020 Aug;51(6):482-491.- DOI: 10.1016/j.arcmed.2020.05.012.
3) Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein.
Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, Hajduk P, Mack J, Fesik SW, Olejniczak ET.
Biochemistry. 2004 May 25;43(20):6059-63 - DOI: 10.1021/bi036155b.
4) SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism.
Lu X, Pan J, Tao J, Guo D.
Virus Genes. 2011 Feb;42(1):37-45.- DOI: 10.1007/s11262-010-0544-x.
5) The nucleocapsid protein of SARS-associated coronavirus inhibits B23 phosphorylation.
Zeng Y, Ye L, Zhu S, Zheng H, Zhao P, Cai W, Su L, She Y, Wu Z..
Biochem Biophys Res Commun. 2008 May 2;369(2):287-91.- DOI: 10.1016/j.bbrc.2008.01.096..
6) The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells.
Surjit M, Liu B, Chow VT, Lal SK.
J Biol Chem. 2006 Apr 21;281(16):10669-81.- DOI: 10.1074/jbc.M509233200.
7) A structural analysis of M protein in coronavirus assembly and morphology.
Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG, Stamou DG, Wilson IA, Kuhn P, Buchmeier MJ.
J Struct Biol. 2011 Apr;174(1):11-22.- DOI: 10.1016/j.jsb.2010.11.021
8) The membrane protein of SARS-CoV suppresses NF-kappaB activation.
Fang X, Gao J, Zheng H, Li B, Kong L, Zhang Y, Wang W, Zeng Y, Ye L.
J Med Virol. 2007 Oct;79(10):1431-9.- DOI: 10.1002/jmv.20953.
9) Conserved HLA binding peptides from five non-structural proteins of SARS-CoV-2-An in silico glance.
Marchan J.
Hum Immunol. 2020 Aug 13:S0198-8859(20)30377-3.- DOI: 10.1016/j.humimm.2020.08.001.
10) Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells.
Narayanan K, Huang C, Lokugamage K, Kamitani W, Ikegami T, Tseng CT, Makino S.
J Virol. 2008 May;82(9):4471-9.- DOI: 10.1128/JVI.02472-07.
11) Severe acute respiratory syndrome coronavirus nonstructural protein 2 interacts with a host protein complex involved in mitochondrial biogenesis and intracellular signaling..
Cornillez-Ty CT, Liao L, Yates JR 3rd, Kuhn P, Buchmeier MJ.
J Virol. 2009;83(19):10314-10318.- DOI: 10.1128/JVI.00842-09
12) Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease.
Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z.
J Gen Virol. 2014 Mar;95(Pt 3):614-626.- DOI: 10.1099/vir.0.059014-0
13) Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles.
Angelini MM, Akhlaghpour M, Neuman BW, Buchmeier MJ.
mBio. 2013 Aug 13;4(4):e00524-13.- DOI: 10.1128/mBio.00524-13.
14) A SARS-CoV-2-Human Protein-Protein Interaction Map Reveals Drug Targets and Potential Drug-Repurposing.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, O'Meara MJ, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Naing ZZC, Zhou Y, Peng S, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Shen W, Shi Y, Zhang Z, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Ramachandran R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Lin Y, Wankowicz SA, Bohn M, Trenker R, Young JM, Cavero D, Hiatt J, Roth T, Rathore U, Subramanian A, Noack J, Hubert M, Roesch F, Vallet T, Meyer B, White KM, Miorin L, Agard D, Emerman M, Ruggero D, García-Sastre A, Jura N, von Zastrow M, Taunton J, Schwartz O, Vignuzzi M, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor S, Fraser JS, Gross J, Sali A, Kortemme T, Beltrao P, Shokat K, Shoichet BK, Krogan NJ.
bioRxiv [Preprint]. 2020 Mar 22:2020.03.22.002386.- DOI: 10.1101/2020.03.22.002386.
15) The Proteins of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS CoV-2 or n-COV19), the Cause of COVID-19.
Yoshimoto FK.
Protein J. 2020 Jun;39(3):198-216.- DOI: 10.1007/s10930-020-09901-4.
16) The SARS-coronavirus nsp7+nsp8 complex is a unique multimeric RNA polymerase capable of both de novo initiation and primer extension.
te Velthuis AJ, van den Worm SH, Snijder EJ.
Nucleic Acids Res. 2012 Feb;40(4):1737-47.- DOI: 10.1093/nar/gkr893.
17) Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Littler DR, Gully BS, Colson RN, Rossjohn J.
iScience. 2020 Jul 24;23(7):101258.- DOI: 10.1016/j.isci.2020.101258.
18) The crystal structure of nsp10-nsp16 heterodimer from SARS-CoV-2 in complex with S-adenosylmethionine.
Rosas-Lemus M, Minasov G, Shuvalova L, Inniss NL, Kiryukhina O, Wiersum G, Kim Y, Jedrzejczak R, Maltseva NI, Endres M, Jaroszewski L, Godzik A, Joachimiak A, Satchell KJF.
bioRxiv [Preprint]. 2020 Apr 26:2020.04.17.047498.- DOI: 10.1101/2020.04.17.047498.
19) SARS-CoV-2 and ORF3a: Nonsynonymous Mutations, Functional Domains, and Viral Pathogenesis.
Issa E, Merhi G, Panossian B, Salloum T, Tokajian S.
mSystems. 2020 May 5;5(3):e00266-20.- DOI: 10.1128/mSystems.00266-20.
20) SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant.
Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ; USFQ-COVID19 Consortium, Nakagawa S, Sato K.
Cell Rep. 2020 Sep 22;32(12):108185.- DOI: 10.1016/j.celrep.2020.108185.
21) Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists.
Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P.
J Virol. 2007 Jan;81(2):548-57.- DOI: 10.1128/JVI.01782-06.
22) Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane.
Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS.
J Virol. 2007 Sep;81(18):9812-24.- DOI: 10.1128/JVI.01012-07.
23) Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference.
Taylor JK, Coleman CM, Postel S, Sisk JM, Bernbaum JG, Venkataraman T, Sundberg EJ, Frieman MB.
J Virol. 2015 Dec;89(23):11820-33.- DOI: 10.1128/JVI.02274-15
24) The ORF7b protein of severe acute respiratory syndrome coronavirus (SARS-CoV) is expressed in virus-infected cells and incorporated into SARS-CoV particles.
Schaecher SR, Mackenzie JM, Pekosz A.
J Virol. 2007 Jan;81(2):718-31.- DOI: 10.1128/JVI.01691-06.
25) Discovery and Genomic Characterization of a 382-Nucleotide Deletion in ORF7b and ORF8 during the Early Evolution of SARS-CoV-2.
Su YCF, Anderson DE, Young BE, Linster M, Zhu F, Jayakumar J, Zhuang Y, Kalimuddin S, Low JGH, Tan CW, Chia WN, Mak TM, Octavia S, Chavatte JM, Lee RTC, Pada S, Tan SY, Sun L, Yan GZ, Maurer-Stroh S, Mendenhall IH, Leo YS, Lye DC, Wang LF, Smith GJD.
mBio. 2020 Jul 21;11(4):e01610-20.- DOI: 10.1128/mBio.01610-20.
26) On the origin and continuing evolution of SARS-CoV-2
Xiaolu Tang, Changcheng Wu, Xiang Li, Yuhe Song, Xinmin Yao, Xinkai Wu, Yuange Duan, Hong Zhang, Yirong Wang, Zhaohui Qian, Jie Cui, and Jian Lu
Natl Sci Rev. 2020;nwaa036.- DOI: 10.1093/nsr/nwaa036
27) The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans
Katarzyna Pancer, Aleksandra Milewska, Katarzyna Owczarek, Agnieszka Dabrowska, Wojciech Branicki, Marek Sanak, Krzysztof Pyrc
bioRxiv 2020.08.29.257360- DOI: 10.1101/2020.08.29.257360