Coronavirus SARS-CoV Related Peptides
The SARS-CoV coronaviruses
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, 2019-nCoV), the coronavirus responsible for coronavirus disease 2019 (COVID-19), shares very high similarity with SARS-CoV.
Both belong to the coronaviruses family, which are enveloped, single-stranded RNA viruses They mainly infect host lung cells through binding to the Angiotensin Converting Enzyme 2 (ACE2) receptor.
Like other coronaviruses, SARS-CoV-2 has four structural proteins: Spike glycoprotein (S), Nucleocapsid (N), Envelope (E), and Membrane (M) proteins.
Spike glycoprotein: a diagnostic and therapeutic target
The Spike glycoprotein mediates the virus attachment to host cell surface receptors and facilitates virus entry by assisting fusion between viral and host cell membranes. It is the most exposed and immunogenic viral protein and hence a target of choice for diagnostic and therapeutic assays.
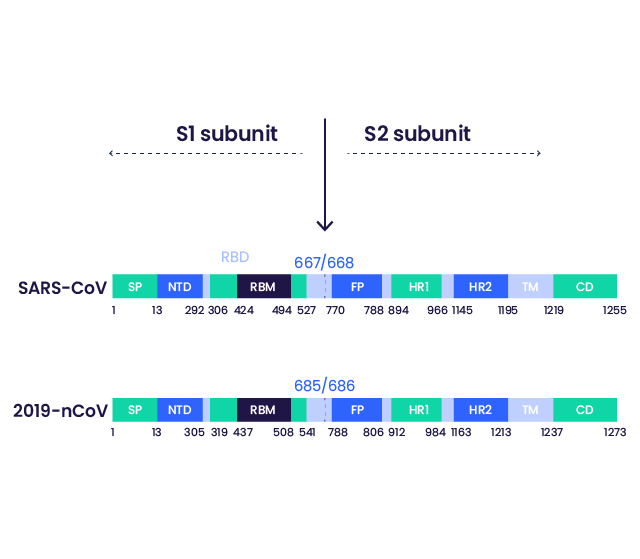
The Spike glycoprotein of SARS-CoV-2 has two furin protease cleavage sites: one at the boundary between S1 and S2 subunits, and one within the S2 subunit:
Figure : Schematic representation of the Spike protein of SARS-CoV and SARS-CoV-2
SP: signal peptide; NTD: N-terminal domain; RBD: receptor binding domain; RBM: receptor binding motif; FP: fusion peptide; HR1: heptad repeat 1; HR2: heptad repeat 2; TM: transmembrane domain; CD: cytoplasmic domain
The S1/S2 cleavage site is indicated.
SARS-CoV proteins as protease targets
The SARS-CoV viral proteins have been identified as targets of several proteases, among which are Furin, Pro-NPY, 3CLpro (3C-like viral protease) and Cathepsins (B, L).
It has been shown that MERS-CoV contains a furin cleavage site just upstream of the fusion peptide. The spike protein of some coronaviruses is cleaved at the S1/S2 boundary by furin(-like) proteases during transport of the newly assembled virions through the secretory pathway. Hence, screening for furin inhibitors observed with furin-dependent viral replication may present as a potential target for drug design.
A peptide substrate custom synthesized by AnaSpec was used to measure the enzymatic activity of 3CLpro in the context of a coronaviral replication mechanism. (Tomar S, et al. 2015).
AnaSpec, the expert of FRET peptide substrates, is offering a range of catalog FRET peptides labeled with our QXL quenchers and fluorescent dyes. Custom sequences can be provided as custom.
FRET peptide based, Protease Activity Assay Kits also available
| SensoLyte® 390 ACE2 Activity Assay Kit *Fluorimetric* | Order Now |
|---|---|
| SensoLyte® 520 Cathepsin B Assay Kit *Fluorimetric* | Order Now |
| SensoLyte® 440 Cathepsin B Assay Kit *Fluorimetric* | Order Now |
| SensoLyte® Rh110 Cathepsin L Assay Kit *Fluorimetric* | Order Now |
| SensoLyte® 520 Cathepsin L Assay Kit *Fluorimetric* | Order Now |
| SensoLyte® Rh110 Furin Activity Assay Kit *Fluorimetric* | Order Now |
Coronavirus-derived peptides
Our expert and high-quality custom synthesis services can provide critical peptide domains/regions or substrates needed to understand coronaviral fusion mechanisms.
Synthesis of peptides such as the Heptad Repeat (HR) regions, which are characteristic of the viral spike proteins could be beneficial in the development of novel therapeutic agents against COVID-19.
Featured Citations
Measurement of Angiotensin Converting Enzyme 2 Activity in Biological Fluid (ACE2)
Xiao F, Burns K.D. (2017)
In: Touyz R., Schiffrin E. (eds) Hypertension. Methods in Molecular Biology, vol 1527. Humana Press, New York, NY.
Identification of a broad-spectrum antiviral small molecule against severe acute respiratory syndrome coronavirus and Ebola, Hendra, and Nipah viruses by using a novel high-throughput screening assay.
Elshabrawy HA, Fan J, Haddad CS, et al.
J Virol. 2014;88(8):4353–4365.
SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection.
Gassen NC, Niemeyer D, Muth D, Corman VM, Martinelli S, Gassen A, Hafner K, Papies J, Mösbauer K, Zellner A, Zannas AS.
Nature Communications. 2019 Dec 18;10(1):1-6.
Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro) IMPLICATIONS FOR nsp5 REGULATION AND THE DEVELOPMENT OF ANTIVIRALS.
Tomar S, Johnston ML, John SE, Osswald HL, Nyalapatla PR, Paul LN, Ghosh AK, Denison MR, Mesecar AD.
Journal of Biological Chemistry. 2015 Aug 7;290(32):19403-22.
Dissecting virus entry: replication-independent analysis of virus binding, internalization, and penetration using minimal complementation of β-galactosidase.
d C, Bloyet LM, Wicht O, et al.
PLoS One. 2014;9(7):e101762.
References
1. Li, W. et al. (2003) Nature 426, 450.
2. Lu R., et al. (2020) Lancet 395, 565–574
3. Du, L., et al. (2009) Nat. Rev. Microbiol 7, 226–236
4. Elshabrawy HA, et al. (2014). Journal of Virology. 88(8), 4353-65.
5. Burkard C, et al. (2014). PLoS One. 2014; 9(7):e101762.